Nanotube coordinate generator with a viewer
for Windows
Japanese Mode is Here
Updated: '04/9/14
With our program you can make Cartesian coordinates
of nanotubes with
specified (n, m) chirality.
The unitcell length or arbitrary length can
be chosed.
Molecular viewer is also included.
The wrapping process of a graphitic sheet
to a nanotube can be animated.
Download the following file:
wrapping.zip (568 KB)
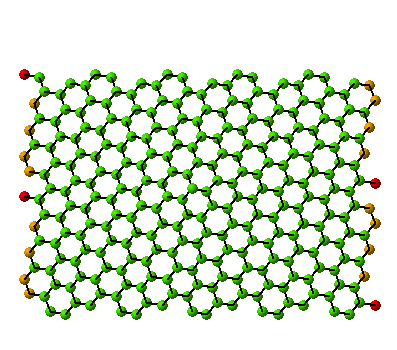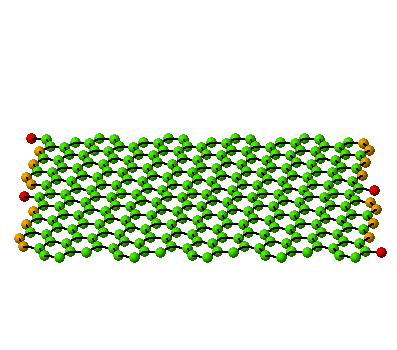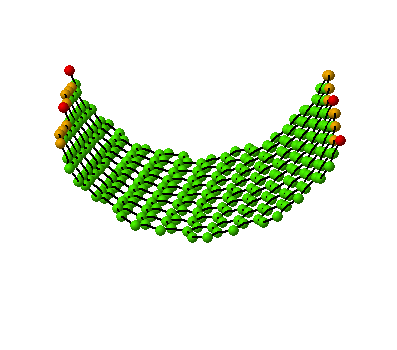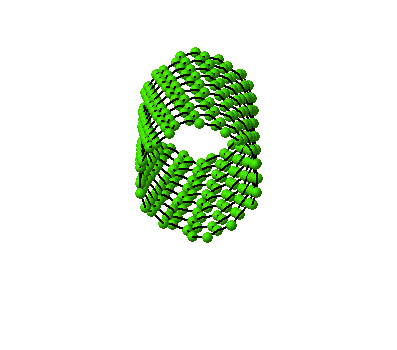 |
(10,5) Nanotube:
tube length is 2xunitcell,
[nseps]=10 |
Quick Usage:
- After downloading wrapping.zip, melt it to
a folder. It is ready to use. No installations
are necessary.
- Run wrapping.exe. Specify chiral index (n,
m), bond length a, tube length, and boundary
length.
The boundary length is only for the visualization
purpose: Specify a larger value or the
same
value as tube length.
Hitting [unitcell] button, the tube length
and boundary length are set as the unitcell
length for your (n, m).
- IMPORTANT NOTE for user of European version of Windows:
In the input line for the bond distance you just have to change the "dot"
to a "comma", if one is using the European windows version.!
Thanks to Prof. Dr. K.-P. Dinse for the advice!
- Other parameters of wrapping.exe (Usually
not necessary to change)
- Numbers for [initial matrix preparation]
specify the internally used initial matrix
size. Usually, default values are
large enough. When using large n or m,
you
may find lacking of nanotube atoms. Then,
modify these numbers to larger
value.
- The value of [Mag] is the magnification factor
for display window of the program. This
display
is prepared for the
understandings of the calculation process
only. So, [Mag] value have no effect
on the
result files.
- By removing the check in [Periodic and only
final shape], intermediate data of wrapping
process are recorded.
By using this the animation can be easily
generated.
- [nsteps] is the number of steps of the wrapping
process.
- Then, push [Make pos.dat] button. After answering
OK to 2 boxes, a ascii data file named
"swnt_pos.dat" is made in the
same
folder as the program. Here, ignoring 7
header
lines,
"swnt_pos.dat" describes x, y,
z Cartesian coordinates with Angstrom units.
- Then, Push [Make pv file] button.
A command line program below is processed.
>hpvc.exe swnt_pos.dat swnt_pos.pv
The ascii file swnt_pos.dat is converted
to a binary file swnt_pos.pv for the
following visualization program.
Depending on your operating system, this
process may not be done. Then,
just use the DOS prompt, change to the
directly
and type the command.
>hpvc.exe swnt_pos.dat swnt_pos.pv
- Then, Push [pvwin] button.
A command line program below is processed.
>pvwin.exe swnt_pos.pv
Now, you can manipulate the visulalization
program.
It is also possible to drag&drop "swnt_pos.pv"
file to "pvwin.exe" program.
- See menus of pvwin.exe for enlargement, rotaion,
animation, Bitmap output,
windows metafile output and more.
"pvwin.exe" is a subset of general
visualization program for molecular dynamics
resutls.
See more details for our homepage.
It is quite powerful program and most of
animations in our animation gallery were produced from this
program.
---Below is a quick instruction to of the
usage of pvwin.exe program--
- Rotation of view: drag with mouse or arrow
keys
- Rotation to an exact angles: Shift-a key
and type alpha and beta value
- Zoom up and down: Ctrl+wheel of mouse, or
+, - keys
- Frame on/off: hit f key
- Animation steps: space key for forward and
backspace key for backward
- Go to a frame number: g key and type frame
number
- Other details: see submenus of view menu
for size of molecule, bond thickness...
- Copy Bitmap file: Ctrl-C or File menu
- Copy Windows meta file: Ctrl-M or File menu
- Save Animation: File menu -> Save Anime:
Successive bitmap files are save at prescribed
folder.
By using some software, you can easily make
animation gif files
Frequently Asked Questions and Answers

